Writing a function in R to solve the valid Sudoku LeetCode problem.
r programming
Longest Common Prefix
solving the longest common prefix LeetCode problem using r
Palindrome Number in R
Palindrome Number LeetCode question using R
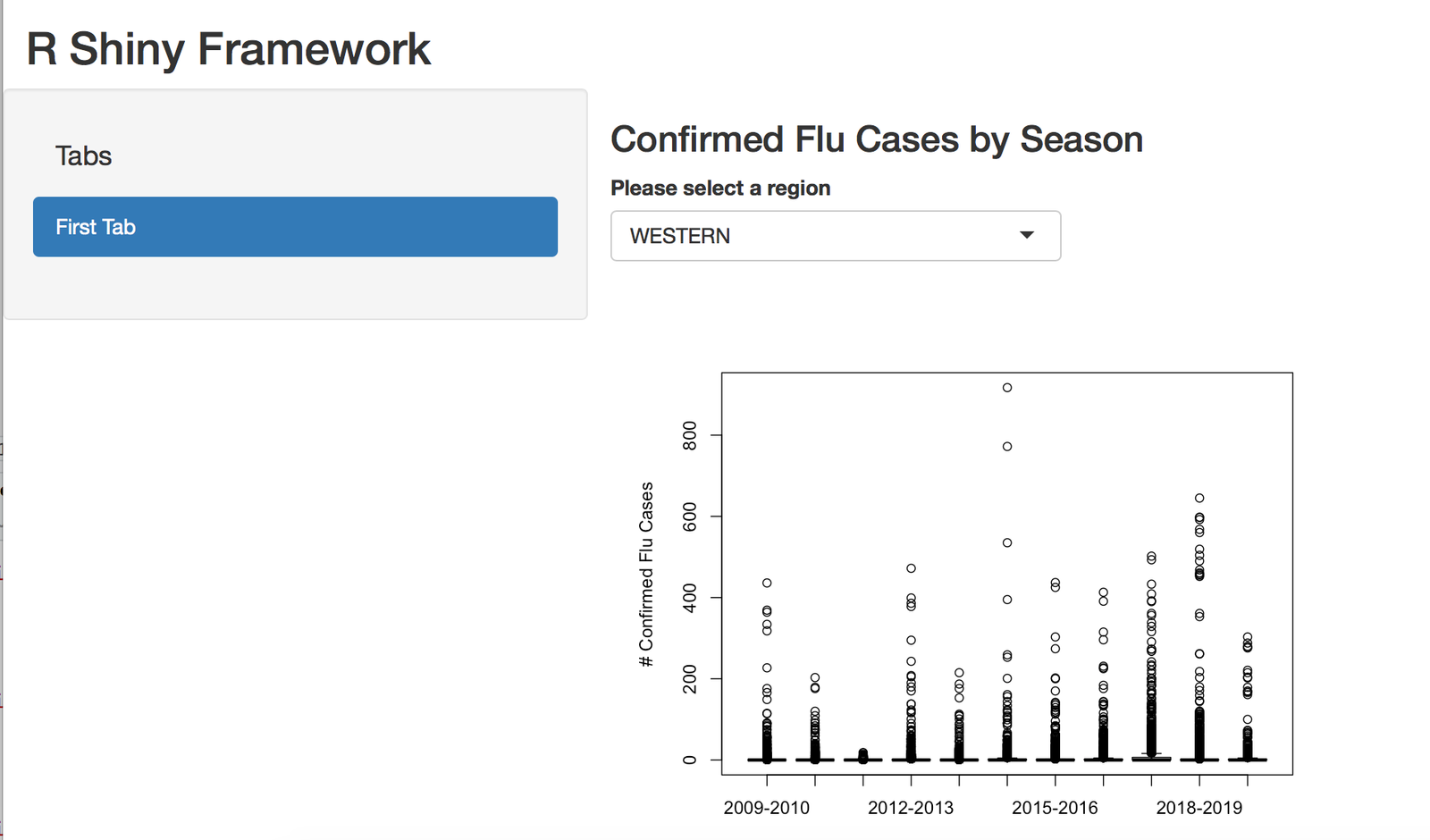
R Shiny Application Split Into Multiple Files
Create an R Shiny Application using multiple files framework to use R Shiny with large applications
Renaming Columns with R
In R you can rename columns multiple ways. You can stick with Base R, use Tidyverse’s dplyr, utilize GREP, and even use data.tables.