A R Shiny application can be completely made of just one file that includes your UI and server code. Depending on the application you’re creating this can get messy as the number of lines grows. Splitting the app across multiple files also helps with debugging and being able to reuse code for another project.
Separating the files for a Shiny Application also helps with version control
For this post, the actual code and what it’s doing will not be covered. The code is more of a placeholder and this is creating a project framework to start Shiny applications.
Creating a new project
The best practice is to create a new project for each application. This is simple with RStudio and using the “File” dropdown to select “New Project”
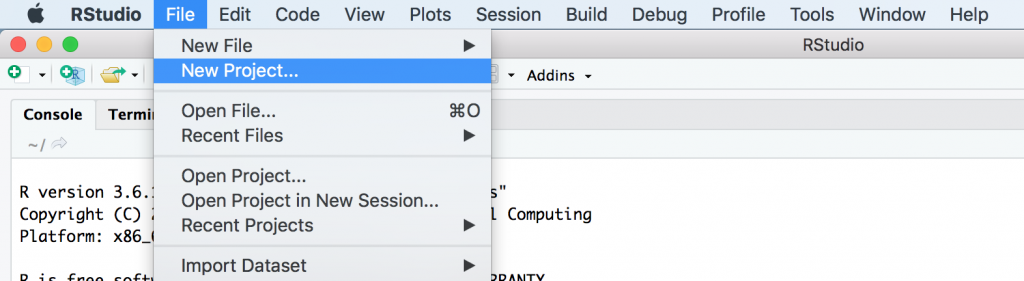



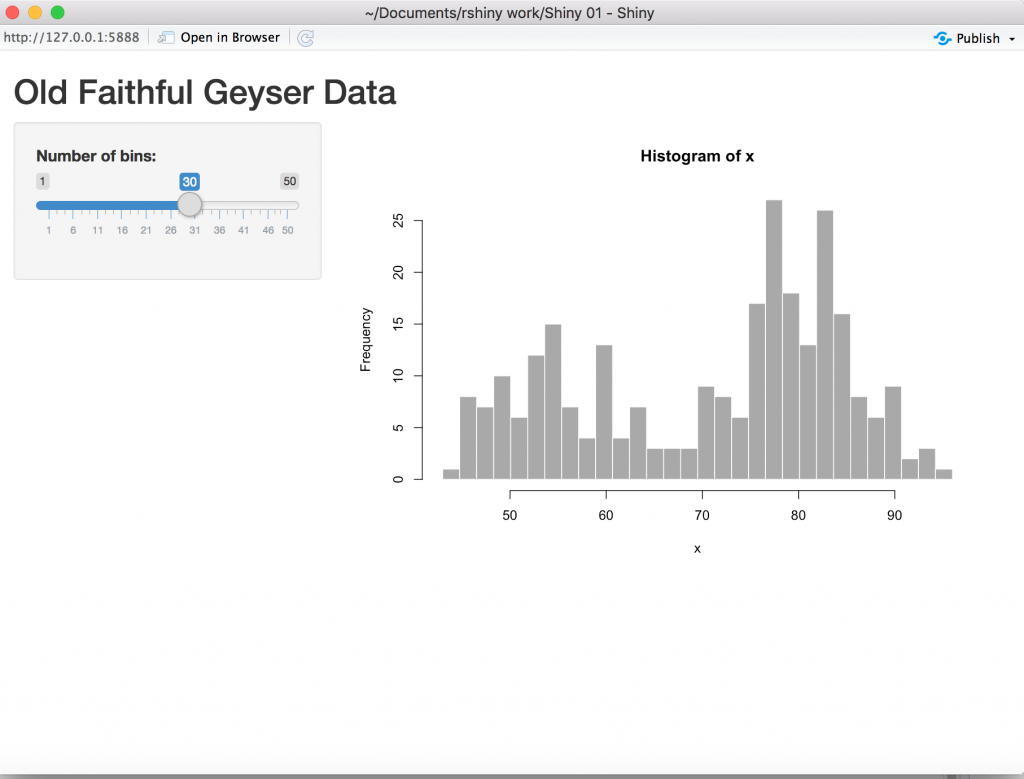
It’s a good idea to run the default Shiny app, if it doesn’t run then you may have installation issues or your security settings do not allow R shiny applications.
Now the code that is used in the example is all in one file, you can delete the file. We will be using 3 files/scripts,
- ui.R
- server.R
- global.R – Not needed but helps to store functions and other code as an application grows large.
I will also create a new subdirectory called data
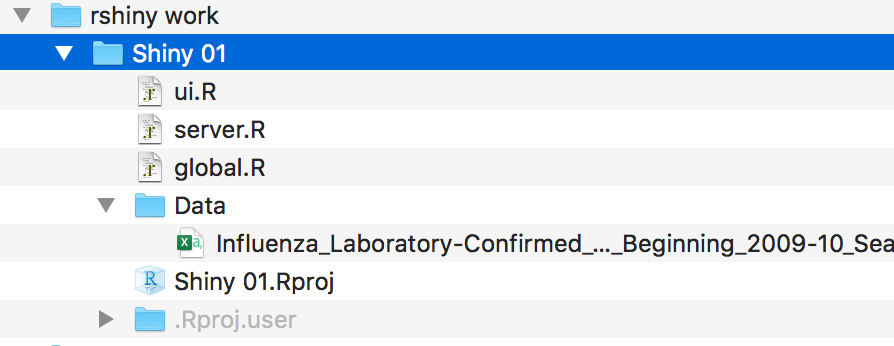
I will also create a new subdirectory called to store the data that will be used.
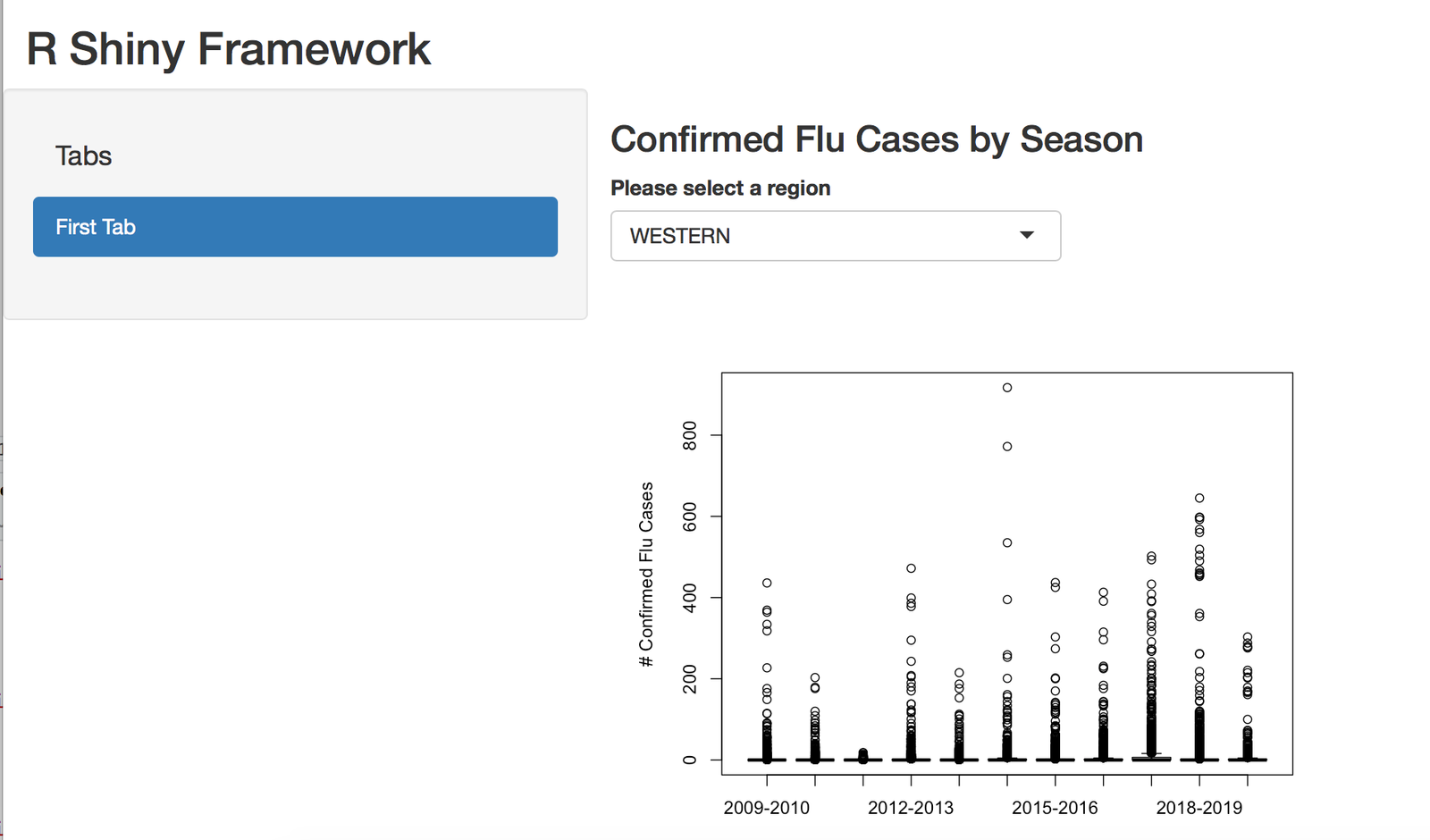
R Shiny Framework Below
UI Code
ui <- fluidPage(
titlePanel("R Shiny Framework"),
navlistPanel(
"Tabs",
tabPanel("First Tab",
h3("Confirmed Flu Cases by Season"),
selectInput("region_id", 'Please select a region',
choices = unique(data$Region),
selected = "WESTERN")
),
mainPanel(
plotOutput(outputId = "plot_id"),
h6("Data Provided by New York State Department of Health"))
)
)Server Script
server <- function(input, output, session) {
output$plot_id <- renderPlot({
temp <- filter(data, Region == input$region_id)
plot(x = temp$Season, y = temp$Count, xlab = "Season", ylab = "# Confirmed Flu Cases")
})
}Global Script
library(shiny)
library(tidyverse)
# Data Provided by New York State Department of Health
# https://health.data.ny.gov/Health/Influenza-Laboratory-Confirmed-Cases-By-County-Beg/jr8b-6gh6
data <- read.csv("./Data/Influenza_Laboratory-Confirmed_Cases_By_County__Beginning_2009-10_Season.csv")
End Results